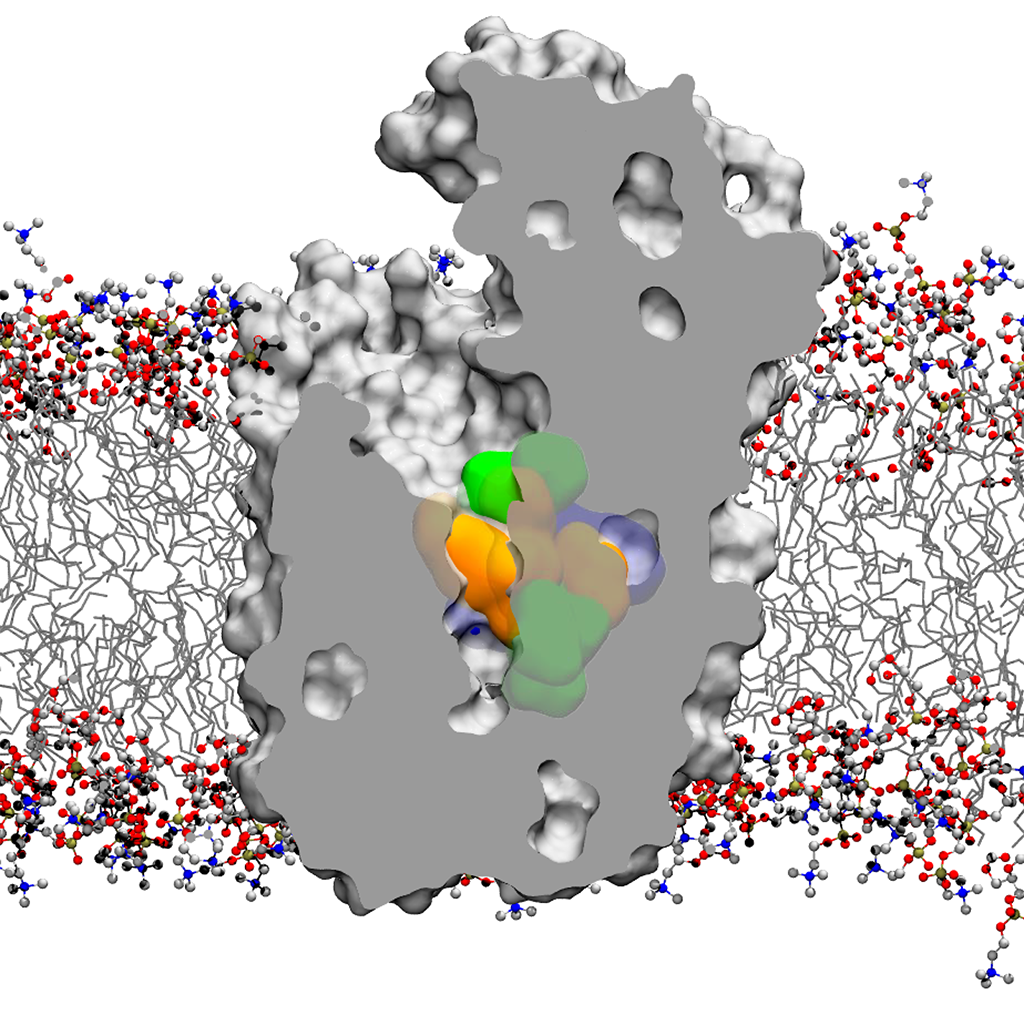
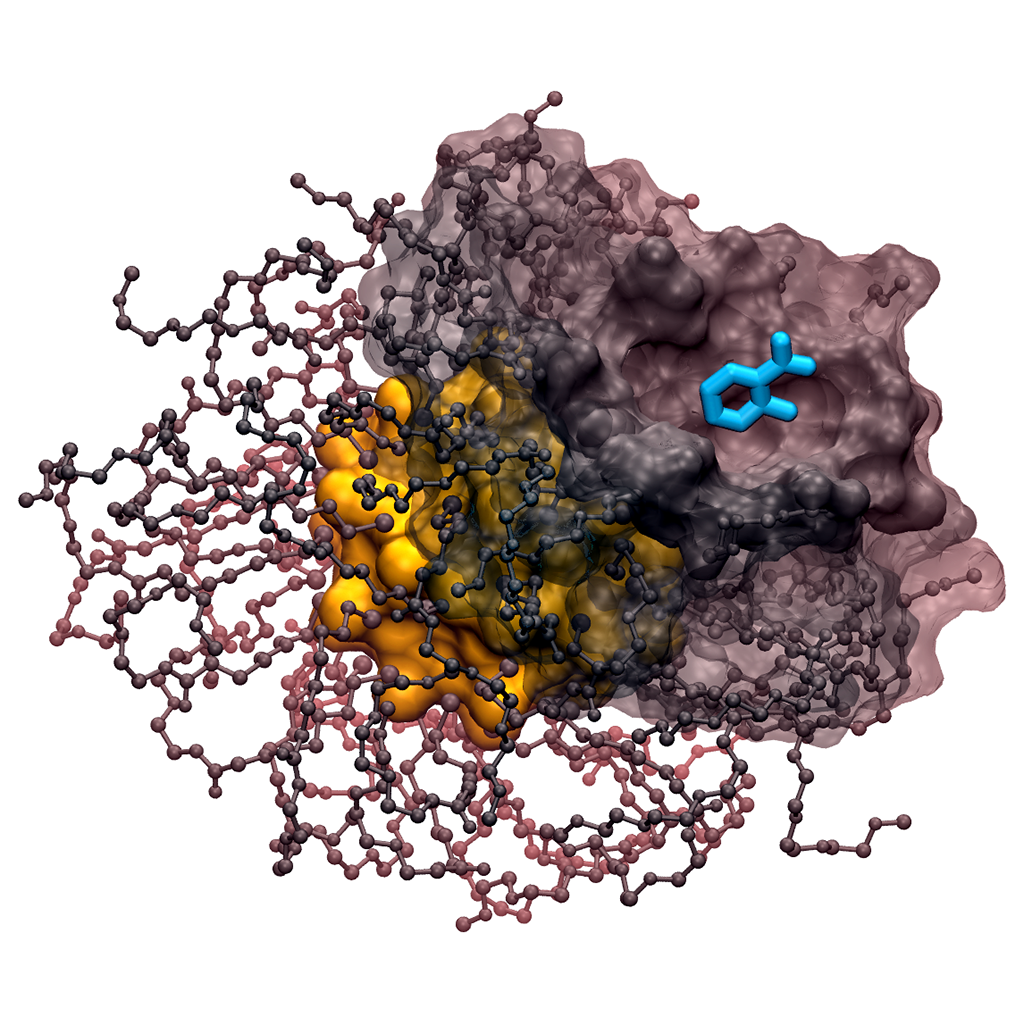
In our group, structure, dynamics and reactivity of the chemical systems of interest are studied by means of classical molecular dynamics (MD) simulations coupled with first-principles-based computational methods (e.g. QM/MM simulations) and enhanced sampling techniques for free energy estimation. This multiscale computational approach includes also docking, virtual screening, de novo small molecules computational design and in silico ADMET evaluation (for drug design). These computational efforts are integrated with medicinal chemistry and structural/molecular biology to identify and optimize novel small molecules able to inhibit the enzymatic function of interest. The goal is to understand the general principles that control molecular recognition and catalysis and use this information to design intelligent nanosystems and potent inhibitors for drug discovery.
Our major focus for drug discovery are targets related to cancer, neurological disorders inflammatory-related diseases.